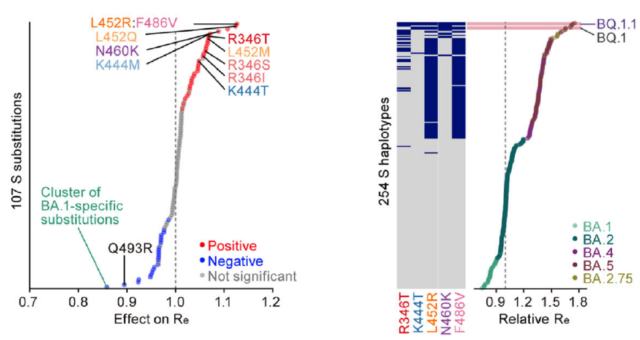
A multi-institute team, including researchers from ICReDD and led by Professor Kei Sato from the Division of Systems Virology at the University of Tokyo’s Institute of Medical Science, has identified mutations that contributed to the increase in viral fitness (the potential to reproduce and spread) during the evolution of the Omicron variant. Evolutionary lineage analysis revealed that various Omicron sublineages have convergently acquired mutations in five important amino acid residues of the spike protein.
Through epidemic modeling analysis, it was shown that the acquisition of these five convergent mutations enhances the fitness of the virus. The modeling results also demonstrated that i) strains with a higher number of convergent mutations exhibit higher fitness, and ii) the acquisition of the five convergent mutations explains a significant portion of the fitness increase during the evolution of the Omicron variant. These findings indicate that the seemingly complex evolution pattern of the Omicron variant can be explained by a simple rule: the more convergent mutations acquired, the higher the fitness.
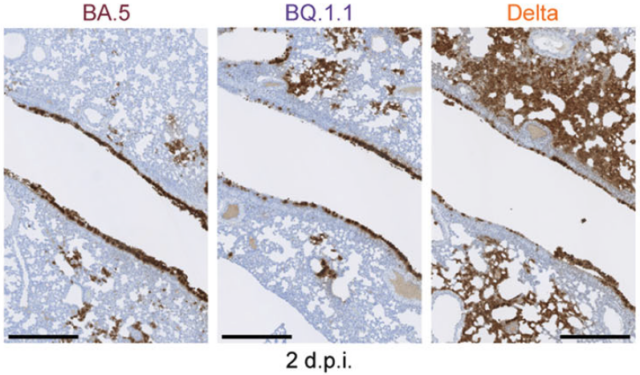
Furthermore, this study conducted detailed virological experiments on the Omicron BQ.1.1 subvariant, which acquired all five convergent mutations and exhibited high fitness. The results revealed that the Omicron BQ.1.1 strain, through the acquisition of convergent mutations, gained higher ACE2 binding affinity, infectivity, and evasion ability against humoral immunity compared to its ancestor strain, Omicron BA.5. However, the pathogenicity of the Omicron BQ.1.1 strain in experimental animal models was found to be similar to that of the Omicron BA.5 strain.
These research findings were published online on May 11, 2023 in Nature Communications.