Keypoints
・Omicron XBB.1.5 virological properties were characterized from multiple perspectives
・Viral proteins and their amino acids involved in changes in viral properties were identified
・These findings contribute to clarifying the full picture of the ecology of the SARS-CoV-2 virus
Research Summary
Collaborative work between the group of Professor Takasuke Fukuhara and Lecturer Tomokazu Tamura at the Graduate School of Medicine, Hokkaido University, and “The Genotype to Phenotype Japan (G2P-Japan)” research consortium, which includes the lab of Professor Shinya Tanaka of ICReDD, revealed the virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant, which emerged around October 2022.
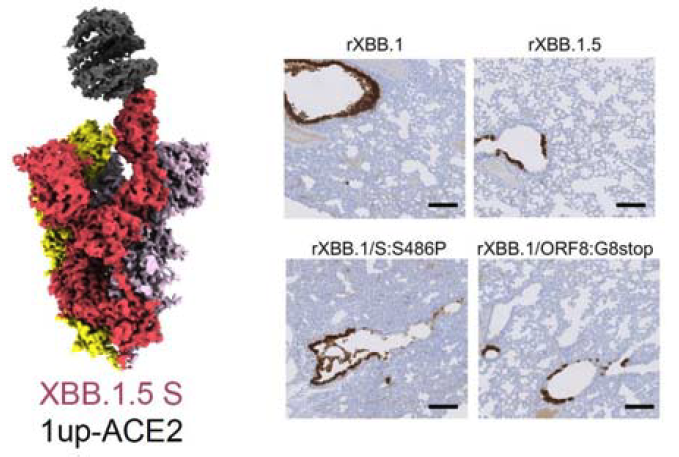
In comparison to the previously prevalent XBB.1, it was known that XBB.1.5 had amino acid point mutations in the spike protein and ORF8 protein. However, it was unclear why the two amino acid differences led to the spread of XBB.1.5. In this study, the characteristics of XBB.1.5 was analyzed from multiple perspectives, including lineage evolution, spike protein structure, and virological properties.
First, phylogenetic analysis of Omicron XBB.1.5 revealed that the mutation in the ORF8 protein appeared first, followed by the mutation in the spike protein. Neutralization assays were then conducted using XBB.1.5 and its direct ancestor XBB.1, showing the two strains have equivalent neutralization susceptibility. Cryogenic electron microscopy analysis did not show differences in the interaction of the spike protein of XBB.1 and XBB.1.5 with the ACE2 receptor, which is the infection receptor for the novel coronavirus. Additionally, no significant differences were observed between XBB.1 and XBB.1.5 in syncytium formation activity and proliferation ability in cultured cells and organoids. However, it became apparent that the pathogenicity of XBB.1.5 in the hamster model was lower than that of XBB.1. To clarify the mechanism of viral attenuation, researchers examined the expression of human MHC class I molecules in organoids. XBB.1 was able to significantly reduce the expression of these molecules, which normally trigger the body’s immune response, but for XBB 1.5 the expression of MHC class I molecules was not suppressed. Pathogenicity tests were performed using recombinant viruses with each point mutation, and it was found that the decrease in pathogenicity of XBB.1.5 was associated with the impairment of the immunosuppressive mechanism due to the loss of ORF8 protein function.
This work was published in the journal Nature Communications on February 8th, 2024.